 We are aiming to establish a ZF-MODELS database in order to fulfill three needs of the project:
We are aiming to establish a ZF-MODELS database in order to fulfill three needs of the project:
- To complement ZFIN, the NIH-funded database of the zebrafish community, by providing a worldwide repository for types of data not currently represented in ZFIN, in particular quantitative data from expression profiling experiments, which will account for the great majority of the data volume and bioinformatics effort.
- To provide a unified user interface for all types of data from our Integrated Project and to distribute raw and processed data to consortium members as well as to scientific and industrial users outside the consortium. In this way the database will fulfill a vital function for the integration of our efforts.
- To support novel ways of data mining by integrating different types of data our database. 2D, 3D or 4D (time lapse) images of GFP expression will be linked to quantitative expression data obtained from the corresponding cell types. These reconstructions will focus particularly on the brain, since an improved understanding of brain disease is a central scientific aim of our project. Furthermore, expression profiles and proteomics data for specific genes in a multitude of conditions will be linked to the annotated genome sequence generated by Ensembl, and to images of in situ expression patterns stored in ZFIN, providing a unique view of gene expression in a developing vertebrate.
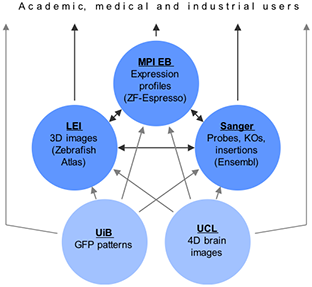
The consortium has decided to pursue a distributed approach for the implementation of the project database. This approach involves several of the partners: MPI EB hosts expression profiling data, Sanger descriptions of oligonucleotide probes, LEI 3D images of zebrafish development, and UiB images of GFP expression patterns. Three different entry points to the data are provided: By MPI EB from expression profiling results, by Sanger from the Ensembl genome assembly, and by LEI from the already existing 3D atlas. Each of these entry points aims to provide links to all the data sources as well as appropriate links to ZFIN. From the third project year onward, UCL will additionally make available 3D as well as 4D reconstructions of the zebrafish brain. Descriptions of ENU and knock-out mutants as well as images of in situ expression patterns are directly submitted to ZFIN.